This document presents to you basic functionality offered by
cohortBuilder package. You’ll learn here about Source and
Cohort objects, how to configure them with filters and filtering steps.
Later on, we’ll present most common Cohort methods that allow to
manipulate the object and extract useful information about Cohort data
and state.
cohortBuilder vs. dplyr
If you’re familiar with dplyr (or any other data
manipulation package) you may be wondering what
cohortBuilder has been created for.
Our main goal for creating cohortBuilder was to provide
a common syntax for operating (filtering) on any data source you need.
This follows the idea for having dplyr and its database
counterpart dbplyr package.
In order to achieve the goal, we put an emphasis on possibility to
write custom extensions in terms of data source type, or operating
backend (underneath cohortBuilder uses dplyr
to operate on data frames, but you may create an extension using
e.g. data.table). See
vignette("custom-extensions").
The second goal was integration of cohortBuilder with
shiny. The GUI for cohortBuilder is provided
by shinyCohortBuilder package. With this extension you may
easily open Cohort configuration panel locally, or include it in you
custom dashboard.
Data: librarian
To present cohortBuilder’s functionality we’ll be
operating on librarian dataset. librarian is a
list of four tables, storing a sample of book library management
database.
cohortBuilder::librarian
#> $books
#> # A tibble: 17 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-385-50420-9 The Da Vinci Code Crim… Transwor… Dan B… 7
#> 2 0-7679-0817-1 A Short History of Nearly Eve… Popu… Transwor… Bill … 4
#> 3 978-0-15-602943-8 The Time Traveler's Wife Gene… Random H… Audre… 2
#> 4 0-224-06252-2 Atonement Gene… Random H… Ian M… 8
#> 5 0-676-97376-0 Life of Pi Gene… Canongate Yann … 11
#> # ℹ 12 more rows
#>
#> $borrowers
#> # A tibble: 20 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000002 2002-08-10 8196 Windsor Road Muscatine, IA … Ms. … 919-530-5272 standa…
#> 3 000003 2003-02-15 6 Wood Lane Calumet City, IL 604… Inga… 706-669-5694 NA
#> 4 000004 2004-06-14 18 Nut Swamp Road Merrimack, NH … Keys… 746-328-6598 standa…
#> 5 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> # ℹ 15 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rowsTo learn more check ?librarian.
Source object
Every time you work with cohortBuilder the crucial part
is to properly define the data source with set_source
function. Source is an R6 object storing metadata about data and its
origin. The metadata allows cohortBuilder to distinct what
methods to use when performing operations on it.
To define a new source you need to provide data (connection).
Let’s create now a new source storing librarian data. To
do so, we pass one obligatory parameter dtconn to
set_source method.
dtconn stores data connection responsible for informing
cohortBuilder on what data are we gonna work (and what
extension to use, if any).
If you want to operate on R-loaded list of tables, provide
tblist class object. tblist is just a named
list of data frames having tblist class.
Note. In order to create ‘tblist’ object use
tblist, e.g. tblist(mtcars, iris).
Note. In order to convert list of data frames to
‘tblist’ just use as.tblist.
str(as.tblist(librarian), max.level = 1L)
#> List of 4
#> $ books : tibble [17 × 6] (S3: tbl_df/tbl/data.frame)
#> $ borrowers: tibble [20 × 6] (S3: tbl_df/tbl/data.frame)
#> $ issues : tibble [50 × 4] (S3: tbl_df/tbl/data.frame)
#> $ returns : tibble [30 × 2] (S3: tbl_df/tbl/data.frame)
#> - attr(*, "class")= chr "tblist"Let’s proceed with creating the source:
librarian_source <- set_source(
as.tblist(librarian)
)
class(librarian_source)
#> [1] "tblist" "Source" "R6"To learn more about set_source’s arguments check
?set_source.
Cohort object
When Source object is ready, the next step is to create
a Cohort object. Cohort is again an R6 object,
providing methods for operating on data included in
Source.
Cohort is responsible in particular for:
- storing definitions of filters (and filtering steps),
- running filtering and keeping result of it,
- computing and caching filter and data statistics,
- keeping and updating filtering configuration state.
In the standard workflow we build Cohort on top of
Source. We achieve it with cohort
function:
With the existing Cohort we may get underlying data with
get_data:
get_data(librarian_cohort)
#> $books
#> # A tibble: 17 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-385-50420-9 The Da Vinci Code Crim… Transwor… Dan B… 7
#> 2 0-7679-0817-1 A Short History of Nearly Eve… Popu… Transwor… Bill … 4
#> 3 978-0-15-602943-8 The Time Traveler's Wife Gene… Random H… Audre… 2
#> 4 0-224-06252-2 Atonement Gene… Random H… Ian M… 8
#> 5 0-676-97376-0 Life of Pi Gene… Canongate Yann … 11
#> # ℹ 12 more rows
#>
#> $borrowers
#> # A tibble: 20 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000002 2002-08-10 8196 Windsor Road Muscatine, IA … Ms. … 919-530-5272 standa…
#> 3 000003 2003-02-15 6 Wood Lane Calumet City, IL 604… Inga… 706-669-5694 NA
#> 4 000004 2004-06-14 18 Nut Swamp Road Merrimack, NH … Keys… 746-328-6598 standa…
#> 5 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> # ℹ 15 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)We’ll present more methods in the next sections.
Configuring and running filters
The next step in cohortBuilder workflow is configuration
of filters. Filters are responsible for providing necessary logic for
performing related data filtering.
The extensive description of filters can be found at
vignette("custom-filters").
The current version of cohortBuilder provides five types
of build-in filters:
- discrete - return values (in column) matching provided set,
- discrete_text - return values based on provided comma separated values,
- range - return values within the provided range,
- date_range - range version for Date type data,
- multi_discrete - extended version of discrete filter working with multiple conditions.
Let’s define discrete filter that will subset books
table listing books written by Dan Brown.
To do so, we have to define the following parameters calling
filter function:
-
type- type of the filter (one of the above), -
dataset- name of the dataset to apply the filter to, -
variable- name of the variable indatasetto apply the filter to, -
value- vector of values to be applied in filter.
So in our case:
author_filter <- filter(
"discrete",
dataset = "books",
variable = "author",
value = "Dan Brown"
)In order to add the filter to existing Cohort we may use
add_filter method:
librarian_cohort <- librarian_cohort %>%
add_filter(author_filter)Alternatively we may use %->% operator that calls
add_filter underneath:
librarian_cohort <- librarian_cohort %->%
author_filterOr define the filter while creating Cohort:
There are much more options for defining filters. To learn more check
vignette("cohort-configuration").
Note. Cohort is an R6 object, so you may skip reassignment above.
For example:
librarian_cohort %>%
add_filter(author_filter)will also work.
Note. To verify if the filter was configured properly just run:
sum_up(librarian_cohort)
#> >> Step ID: 1
#> -> Filter ID: EEXOM1772118519948
#> Filter Type: discrete
#> Filter Parameters:
#> dataset: books
#> variable: author
#> value: Dan Brown
#> keep_na: TRUE
#> description:
#> active: TRUEThe output highlights list of configured filters along with their
parameters. You can see here the id attached to filter and some extra
parameters such as keep_na or active which we
describe in the next sections.
More to that we can realize the filter was defined in the step with
ID equals to 1. That’s because cohortBuilder allows to
perform multi-stage filtering.
Let’s get back to filtering the books. Configuring
filters only adds proper metadata in the Cohort object, which means data
filtering is not performed automatically. This allows to set the proper
configuration first, and run calculation only once.
If you want to run data filtering, just call run:
run(librarian_cohort)Let’s check if the operation worked fine by checking the resulting data:
get_data(librarian_cohort)
#> $books
#> # A tibble: 2 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adv… Transwor… Dan B… 7
#> 2 0-671-02735-2 Angels and Demons Crime, Thriller & Adv… Transwor… Dan B… 4
#>
#> $borrowers
#> # A tibble: 20 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000002 2002-08-10 8196 Windsor Road Muscatine, IA … Ms. … 919-530-5272 standa…
#> 3 000003 2003-02-15 6 Wood Lane Calumet City, IL 604… Inga… 706-669-5694 NA
#> 4 000004 2004-06-14 18 Nut Swamp Road Merrimack, NH … Keys… 746-328-6598 standa…
#> 5 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> # ℹ 15 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)If you want to run data filtering automatically when the filter is
defined you can set run_flow = TRUE:
librarian_cohort <- librarian_source %>%
cohort() %>%
add_filter(author_filter, run_flow = TRUE)when using add_filter or:
when configuring filter along with creating cohort.
Now when the data filtered, how can we get data state before
filtering? With get_data it’s easy, just set
state = "pre":
get_data(librarian_cohort, state = "pre")
#> $books
#> # A tibble: 17 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-385-50420-9 The Da Vinci Code Crim… Transwor… Dan B… 7
#> 2 0-7679-0817-1 A Short History of Nearly Eve… Popu… Transwor… Bill … 4
#> 3 978-0-15-602943-8 The Time Traveler's Wife Gene… Random H… Audre… 2
#> 4 0-224-06252-2 Atonement Gene… Random H… Ian M… 8
#> 5 0-676-97376-0 Life of Pi Gene… Canongate Yann … 11
#> # ℹ 12 more rows
#>
#> $borrowers
#> # A tibble: 20 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000002 2002-08-10 8196 Windsor Road Muscatine, IA … Ms. … 919-530-5272 standa…
#> 3 000003 2003-02-15 6 Wood Lane Calumet City, IL 604… Inga… 706-669-5694 NA
#> 4 000004 2004-06-14 18 Nut Swamp Road Merrimack, NH … Keys… 746-328-6598 standa…
#> 5 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> # ℹ 15 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)Multi-stage filtering
With cohortBuilder you can define filters in groups
named ‘steps’ or ‘filtering steps’.
Filtering steps allow you to sequentially perform groups of filtering
operations. In order to define step, just wrap set of filters in
step function.
We will define three filters:
- Taking all the books written by Dan Brown.
- Filtering only the members (borrowers) with “standard” program.
- Taking only the books with less than 5 copies.
We’ll include filters 1. and 2. in the first step - filter 3. in the second one.
The below code does the job:
librarian_cohort <- librarian_source %>%
cohort(
step(
filter(
"discrete",
id = "author", dataset = "books",
variable = "author", value = "Dan Brown"
),
filter(
"discrete",
id = "program", dataset = "borrowers",
variable = "program", value = "premium", keep_na = FALSE
)
),
step(
filter(
"range",
id = "copies", dataset = "books",
variable = "copies", range = c(-Inf, 5L)
)
)
)Let’s note a few parts that occurred above:
- For each filter we defined
idparameter. This assigns provided id to each filter what makes accessing it later much easier. - For ‘program’ filter we set
keep_na = FALSEwhat results with excludingNAvalues (the parameter is available for each filter type). - For filtering number of copies we’ve used
rangefilter, for which sub-setting value is defined withrangeparameter.
Let’s check the Cohort configuration:,
sum_up(librarian_cohort)
#> >> Step ID: 1
#> -> Filter ID: author
#> Filter Type: discrete
#> Filter Parameters:
#> dataset: books
#> variable: author
#> value: Dan Brown
#> keep_na: TRUE
#> description:
#> active: TRUE
#> -> Filter ID: program
#> Filter Type: discrete
#> Filter Parameters:
#> dataset: borrowers
#> variable: program
#> value: premium
#> keep_na: FALSE
#> description:
#> active: TRUE
#> >> Step ID: 2
#> -> Filter ID: copies
#> Filter Type: range
#> Filter Parameters:
#> dataset: books
#> variable: copies
#> range: -Inf, 5
#> keep_na: TRUE
#> description:
#> active: TRUEWe can see filters were correctly assigned to each step.
Having multiple steps defined, we can use get_data to
extract resulting data after each step. In order to precise the step we
want to get data from, just pass its id as step_id
parameter:
run(librarian_cohort)
get_data(librarian_cohort, step_id = 1L)
#> $books
#> # A tibble: 2 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adv… Transwor… Dan B… 7
#> 2 0-671-02735-2 Angels and Demons Crime, Thriller & Adv… Transwor… Dan B… 4
#>
#> $borrowers
#> # A tibble: 6 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> 3 000008 2006-11-15 9533 Delaware Dr. Peabody, MA 01… Mrs.… 460-779-8714 premium
#> 4 000011 2009-03-24 745 E. Sussex Drive Mahwah, NJ 0… Mr. … 378-884-6509 premium
#> 5 000013 2011-09-30 534 Iroquois Ave. Watertown, MA … Dr. … 104-832-8013 premium
#> # ℹ 1 more row
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)
get_data(librarian_cohort, step_id = 2L)
#> $books
#> # A tibble: 1 × 6
#> isbn title genre publisher author copies
#> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 0-671-02735-2 Angels and Demons Crime, Thriller & Adv… Transwor… Dan B… 4
#>
#> $borrowers
#> # A tibble: 6 × 6
#> id registered address name phone_number program
#> <chr> <date> <chr> <chr> <chr> <chr>
#> 1 000001 2001-06-09 66 N. Evergreen Ave. Norristown,… Mrs.… 626-594-4729 premium
#> 2 000005 2005-01-15 580 Chapel Rd. Delray Beach, FL … Ferd… 127-363-0738 premium
#> 3 000008 2006-11-15 9533 Delaware Dr. Peabody, MA 01… Mrs.… 460-779-8714 premium
#> 4 000011 2009-03-24 745 E. Sussex Drive Mahwah, NJ 0… Mr. … 378-884-6509 premium
#> 5 000013 2011-09-30 534 Iroquois Ave. Watertown, MA … Dr. … 104-832-8013 premium
#> # ℹ 1 more row
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)Note. When step_id is not provided, the
method returns the last step data.
Note. You may precise if you want to extract data
before or after filtering using state parameter. Because
the proceeding step uses result from the previous one, we have:
Exploring the Cohort object methods
Learning more about the source data
Having Cohort object created, you may want to use its methods for exploring underlying data.
With methods such as:
-
stat, -
plot_data, attrition
you can:
- get filter related data statistics,
- display filter related data on plot,
- display data changes across filtering steps.
stat(librarian_cohort, step_id = 1L, filter_id = "program")
#> $n_data
#> [1] 6
#>
#> $choices
#> $choices$premium
#> [1] 6
#>
#>
#> $n_missing
#> [1] 0
stat(librarian_cohort, step_id = 2L, filter_id = "copies")
#> $n_data
#> [1] 1
#>
#> $frequencies
#> level count l_bound u_bound
#> 1 1 1 4 4
#>
#> $n_missing
#> [1] 0
plot_data(librarian_cohort, step_id = 1L, filter_id = "program")
plot_data(librarian_cohort, step_id = 2L, filter_id = "copies")
attrition(librarian_cohort, dataset = "books")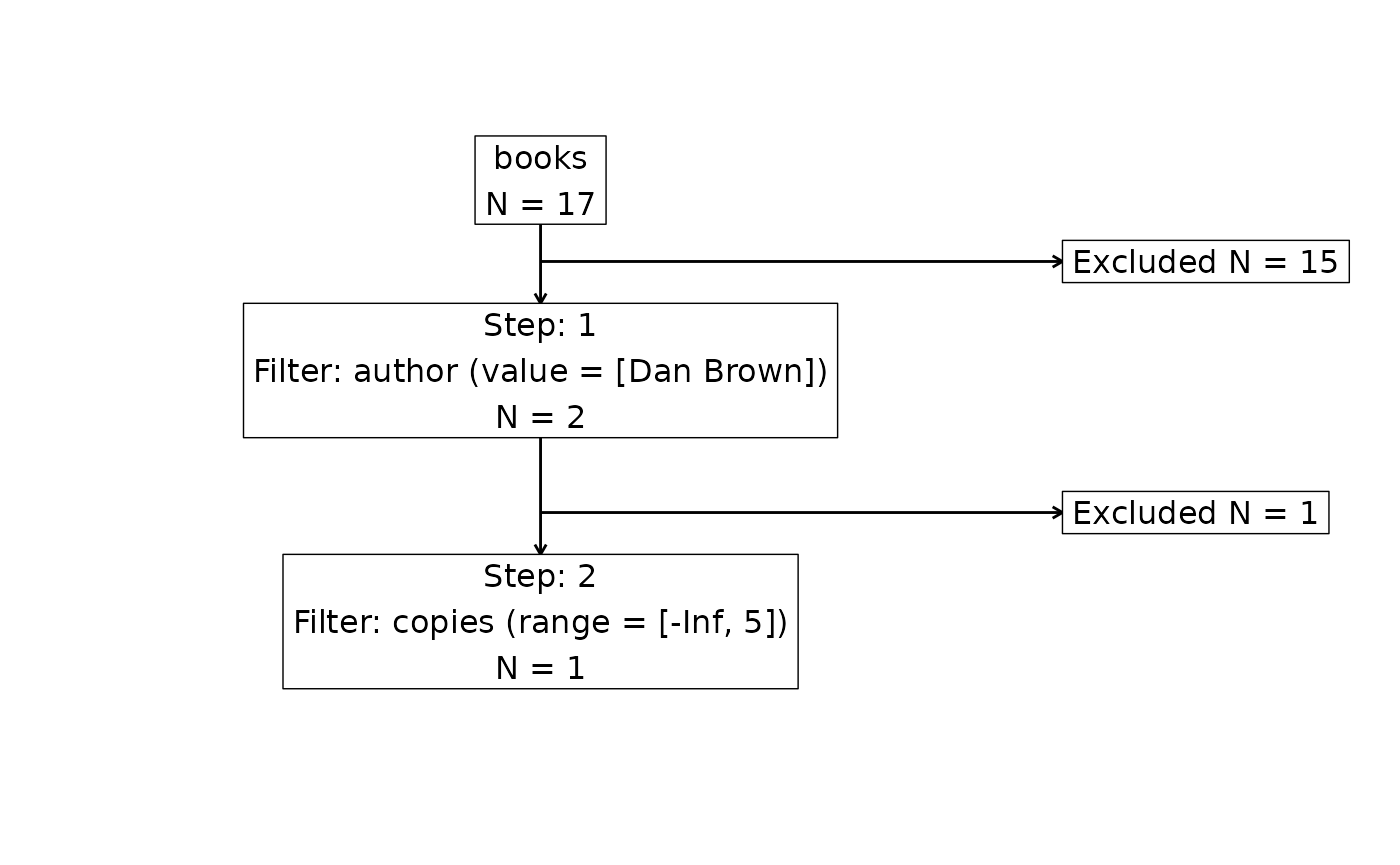
attrition(librarian_cohort, dataset = "borrowers")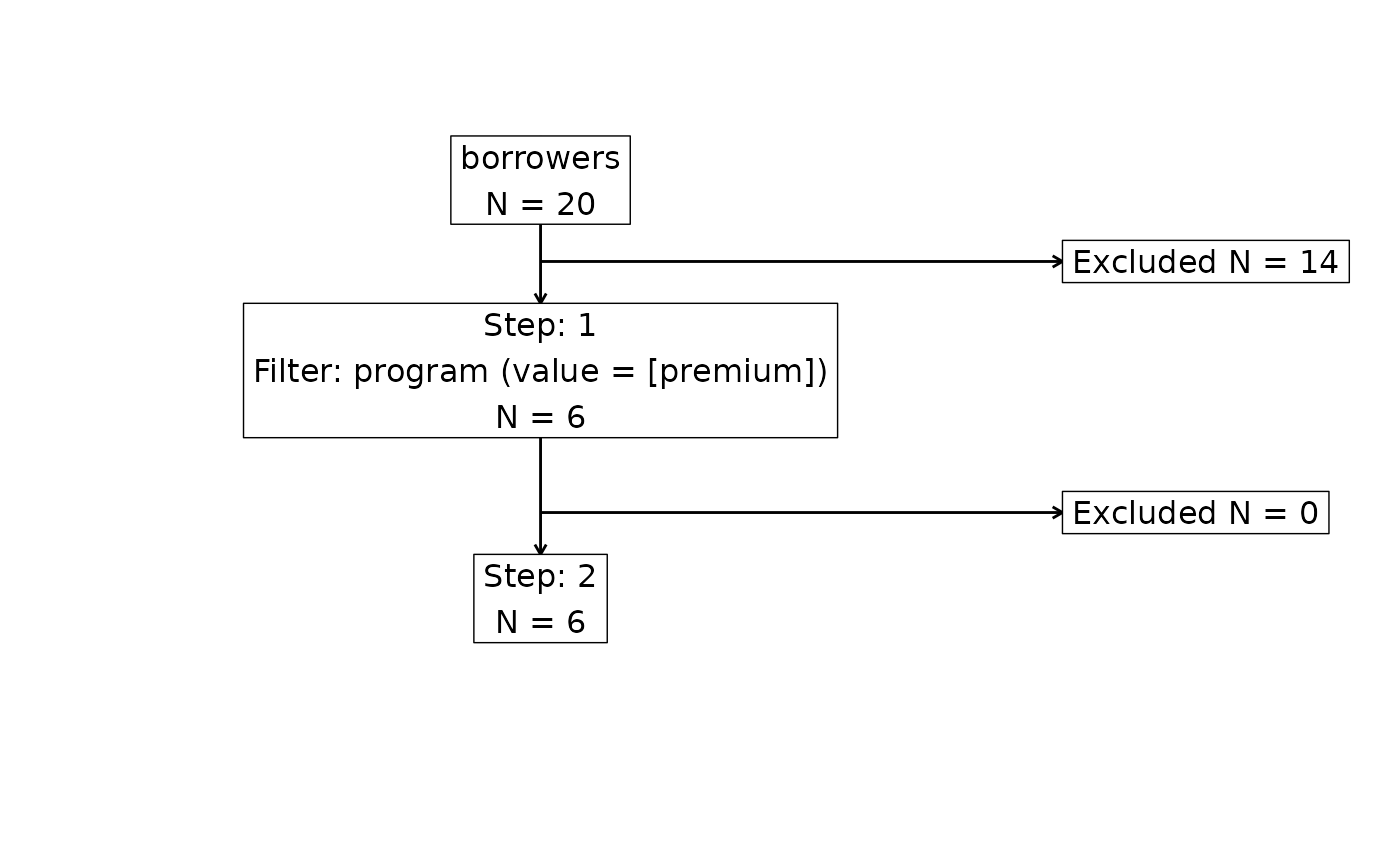
Sharing code and reproducibility
The cohortBuilder package offers some methods to make
sharing the workflow easier.
With code, you may get the reproducible code written
using methods operating on specific source (i.e. dplyr for
tblist and dbplyr for db
source):
code(librarian_cohort)
#> .pre_filtering <- function(source, data_object, step_id) {
#> for (dataset in names(data_object)) {
#> attr(data_object[[dataset]], "filtered") <- FALSE
#> }
#> return(data_object)
#> }
#> .run_binding <- function(source, binding_key, data_object_pre, data_object_post,
#> ...) {
#> binding_dataset <- binding_key$update$dataset
#> dependent_datasets <- names(binding_key$data_keys)
#> active_datasets <- data_object_post %>%
#> purrr::keep(~attr(., "filtered")) %>%
#> names()
#> if (!any(dependent_datasets %in% active_datasets)) {
#> return(data_object_post)
#> }
#> key_values <- NULL
#> common_key_names <- paste0("key_", seq_along(binding_key$data_keys[[1]]$key))
#> for (dependent_dataset in dependent_datasets) {
#> key_names <- binding_key$data_keys[[dependent_dataset]]$key
#> tmp_key_values <- collapse::funique(data_object_post[[dependent_dataset]][,
#> key_names, drop = FALSE]) %>%
#> stats::setNames(common_key_names)
#> if (is.null(key_values)) {
#> key_values <- tmp_key_values
#> } else {
#> key_values <- dplyr::inner_join(key_values, tmp_key_values, by = common_key_names)
#> }
#> }
#> df <- switch(as.character(binding_key$post), `FALSE` = data_object_pre[[binding_dataset]],
#> `TRUE` = data_object_post[[binding_dataset]])
#> data_object_post[[binding_dataset]] <- tryCatch({
#> collapse::join(df, key_values, on = stats::setNames(common_key_names, binding_key$update$key),
#> how = "inner", verbose = getOption("cb_verbose", default = FALSE))
#> }, error = function(e) {
#> dplyr::inner_join(df, key_values, by = stats::setNames(common_key_names,
#> binding_key$update$key))
#> })
#> if (binding_key$activate) {
#> attr(data_object_post[[binding_dataset]], "filtered") <- TRUE
#> }
#> return(data_object_post)
#> }
#> source <- list(dtconn = as.tblist(librarian))
#> data_object <- source$dtconn
#> step_id <- "1"
#> pre_data_object <- data_object
#> data_object <- .pre_filtering(source, data_object, "1")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter(author %in% c("Dan Brown", NA))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object[["borrowers"]] <- data_object[["borrowers"]] %>%
#> dplyr::filter(program %in% "premium")
#> attr(data_object[["borrowers"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "1")
#> for (binding_key in binding_keys) {
#> data_object <- .run_binding(source, binding_key, pre_data_object, data_object)
#> }
#> step_id <- "2"
#> data_object <- .pre_filtering(source, data_object, "2")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter((copies <= 5 & copies >= -Inf) | is.na(copies))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "2")We can see above, the resulting code uses source object,
which creation code can be defined separately while creating it:
librarian_source <- set_source(
as.tblist(librarian),
source_code = quote({
source <- list()
source$dtconn <- as.tblist(librarian)
})
)
librarian_cohort <- librarian_source %>%
cohort(
step(
filter(
"discrete",
id = "author", dataset = "books",
variable = "author", value = "Dan Brown"
),
filter(
"discrete",
id = "program", dataset = "borrowers",
variable = "program", value = "premium", keep_na = FALSE
)
),
step(
filter(
"range",
id = "copies", dataset = "books",
variable = "copies", range = c(-Inf, 5L)
)
),
run_flow = TRUE
)
code(librarian_cohort)
#> .pre_filtering <- function(source, data_object, step_id) {
#> for (dataset in names(data_object)) {
#> attr(data_object[[dataset]], "filtered") <- FALSE
#> }
#> return(data_object)
#> }
#> .run_binding <- function(source, binding_key, data_object_pre, data_object_post,
#> ...) {
#> binding_dataset <- binding_key$update$dataset
#> dependent_datasets <- names(binding_key$data_keys)
#> active_datasets <- data_object_post %>%
#> purrr::keep(~attr(., "filtered")) %>%
#> names()
#> if (!any(dependent_datasets %in% active_datasets)) {
#> return(data_object_post)
#> }
#> key_values <- NULL
#> common_key_names <- paste0("key_", seq_along(binding_key$data_keys[[1]]$key))
#> for (dependent_dataset in dependent_datasets) {
#> key_names <- binding_key$data_keys[[dependent_dataset]]$key
#> tmp_key_values <- collapse::funique(data_object_post[[dependent_dataset]][,
#> key_names, drop = FALSE]) %>%
#> stats::setNames(common_key_names)
#> if (is.null(key_values)) {
#> key_values <- tmp_key_values
#> } else {
#> key_values <- dplyr::inner_join(key_values, tmp_key_values, by = common_key_names)
#> }
#> }
#> df <- switch(as.character(binding_key$post), `FALSE` = data_object_pre[[binding_dataset]],
#> `TRUE` = data_object_post[[binding_dataset]])
#> data_object_post[[binding_dataset]] <- tryCatch({
#> collapse::join(df, key_values, on = stats::setNames(common_key_names, binding_key$update$key),
#> how = "inner", verbose = getOption("cb_verbose", default = FALSE))
#> }, error = function(e) {
#> dplyr::inner_join(df, key_values, by = stats::setNames(common_key_names,
#> binding_key$update$key))
#> })
#> if (binding_key$activate) {
#> attr(data_object_post[[binding_dataset]], "filtered") <- TRUE
#> }
#> return(data_object_post)
#> }
#> source <- list()
#> source$dtconn <- as.tblist(librarian)
#> data_object <- source$dtconn
#> step_id <- "1"
#> pre_data_object <- data_object
#> data_object <- .pre_filtering(source, data_object, "1")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter(author %in% c("Dan Brown", NA))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object[["borrowers"]] <- data_object[["borrowers"]] %>%
#> dplyr::filter(program %in% "premium")
#> attr(data_object[["borrowers"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "1")
#> for (binding_key in binding_keys) {
#> data_object <- .run_binding(source, binding_key, pre_data_object, data_object)
#> }
#> step_id <- "2"
#> data_object <- .pre_filtering(source, data_object, "2")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter((copies <= 5 & copies >= -Inf) | is.na(copies))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "2")What’s more, you can manipulate the output with additional arguments:
-
include_methods- list of methods names which definition should be printed in output, -
include_action- list of actions names (such as “pre_filtering”) that should be included in output, -
modifier- a custom modifier of data.frame storing reproducible code parts, -
mark_step- should step ID be presented in output.
The second option for achieving reproducibility allows to restore cohort configuration using its state. The cohort state is a list (or json) storing information about all the steps and filters configuration.
You may get the state with get_state method:
state <- get_state(librarian_cohort, json = TRUE)
state
#> [{"step":"1","filters":[{"type":"discrete","id":"author","name":"author","variable":"author","value":"Dan Brown","dataset":"books","keep_na":true,"description":null,"active":true},{"type":"discrete","id":"program","name":"program","variable":"program","value":"premium","dataset":"borrowers","keep_na":false,"description":null,"active":true}]},{"step":"2","filters":[{"type":"range","id":"copies","name":"copies","variable":"copies","range":["-Inf",5],"dataset":"books","keep_na":true,"description":null,"active":true}]}]Then, having an empty cohort, use restore to apply the
configuration:
librarian_cohort <- librarian_source %>%
cohort()
restore(librarian_cohort, state = state)
sum_up(librarian_cohort)
#> >> Step ID: 1
#> -> Filter ID: author
#> Filter Type: discrete
#> Filter Parameters:
#> dataset: books
#> variable: author
#> value: Dan Brown
#> keep_na: TRUE
#> description:
#> active: TRUE
#> -> Filter ID: program
#> Filter Type: discrete
#> Filter Parameters:
#> dataset: borrowers
#> variable: program
#> value: premium
#> keep_na: FALSE
#> description:
#> active: TRUE
#> >> Step ID: 2
#> -> Filter ID: copies
#> Filter Type: range
#> Filter Parameters:
#> dataset: books
#> variable: copies
#> range: -Inf, 5
#> keep_na: TRUE
#> description:
#> active: TRUE